Custom-built cryo-fluorescence development microscope
We have constructed a custom cryo-fluorescence microscope stand based on an upright Nikon chassis coupled to a Linkam CMS196 cryo-stage. The sample stage is able to accept standard electron-microscopy grids and discs, in addition to our custom solid-immersion lens assemblies which are capable of boosting the effective numerical aperture of the microscope to beyond that of a commercially available rooom-temperature STORM microscope (see our publication pre-print here).
Lasers
| 405 nm, 488 nm, 561 nm, 642 nm @ suitable power densities for cryo-STORM.
|
Detectors
| Andor iXon EMCCD or Hamamatsu Orca Flash V4.0.
|
Dichroics and filter sets
| We have combinations to allow for imaging most popular flourescent proteins and inorganic dyes.
Multi-colour imaging is achieved sequentially with fudicial registration.
|
Objectives
| 50x 0.55NA Mitutoyo LWD
100x 0.55NA Mitutoyo LWD
454x 2.17NA solid immersion lens
|
Suitable applications
| STORM imaging of bacteria cells, yeast cells, exosomes and drop-cast fluorescently-labelled proteins.
|
Resolution performance
| STORM imaging modality routinely achieves <8 nm localisation precision, offering resolutions <20 nm for optimal samples.
|
Example images from superSIL STORM imaging

Figure 1: SuperSIL resolution characterization and imaging of E. Coli cells under cryogenic conditions. Representative images of sub-diffraction
limited objects from (A) the superSIL microscope and (B) the specialist system. Bar: 0.5 µm.
(C) Box charts of FWHM of the PSFs for
the superSIL and specialist systems
showing the 25th and 75th percentiles of the data points. The whiskers show the
standard deviation. (D) ATP-binding
cassette (ABC) transporter protein PH1735 fused with EGFP in E.coli cells imaged in wide-field superSIL microscopy and (E)
superSIL STORM image. (F) Localization precision histogram
from the image in (E). (G) The enlarged image of the region
of the cell indicated by the red dashed box in (E). (H) The enlarged image of the small region in the
cell indicated by the orange dashed box in (G). (I)
Line profile of the cross-section of two PH1735 protein clusters indicated by
the cyan dashed lines in (H). (J)
Line profile of the cross-section of two PH1735 protein clusters closer to each
other indicated by the red dashed line in (H). (K)
Line profiles of the cross-section of two adjacent single molecules (magenta
and green) indicated by the magenta and green dashed lines in (H).
(L) FRC curve revealing the 12 nm
resolution in the region shown in (H). Bars: 1 μm (D, E), 100 nm (G)
and 20 nm (H).
Depth characteristics of superSILs
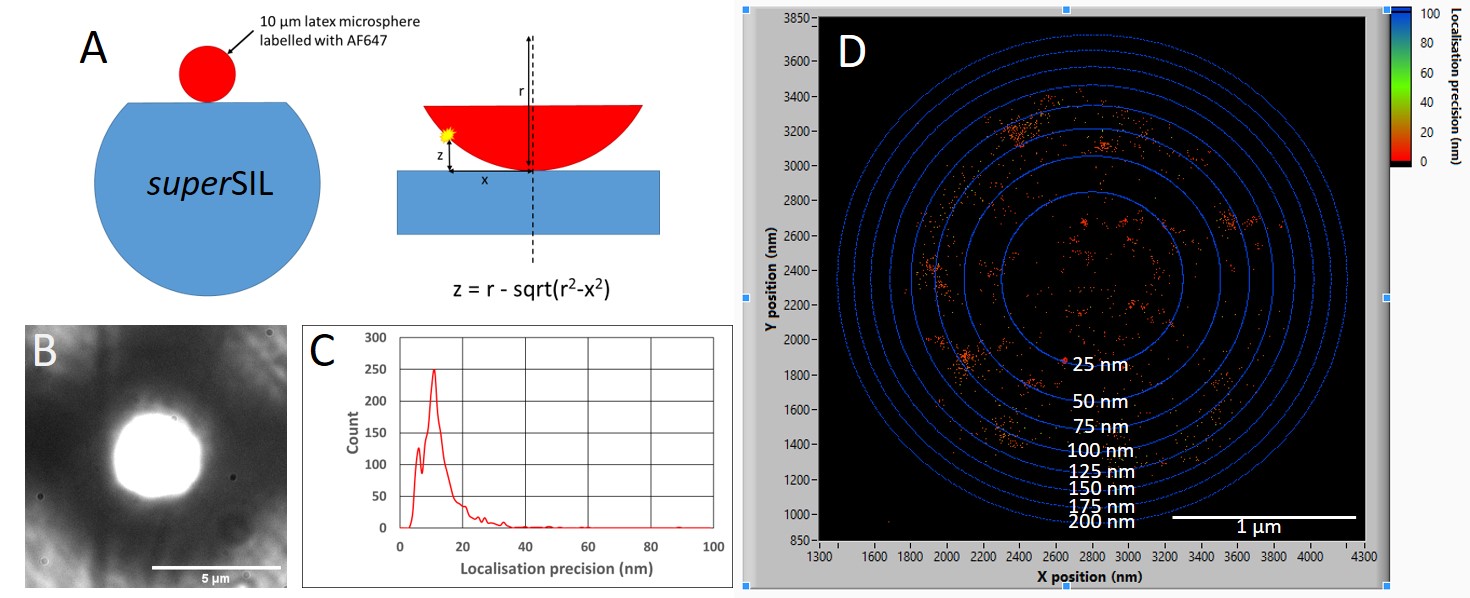
Figure 2:
Characterising the depth of focus STORM performance of a superSIL. A)
10 µm diameter latex microspheres were first functionalised with a biotin
coating and then labelled with streptavidin-AF647 dye. The microspheres were
adhered to a poly-l-lysine coated superSIL and plunge frozen ready for imaging. B)
A brightfield
image of the microsphere shows a bright central focussed region and
out-of-focus periphery. There is a visible scratch on the superSIL face
showing the focal plane of the image is at the surface. A cryo-STORM
measurement was performed and C) shows the a histogram plot revealing a mean
localisation precision of 10 nm. D) An intensity scatter plot shows the
achievable localisation precision is evenly distributed across the in-focus
region of the microsphere. The blue rings surrounding the localisations
indicate calculated distance intervals away from the surface of the superSIL,
revealing high localisation precisions up to a depth of 100 nm.
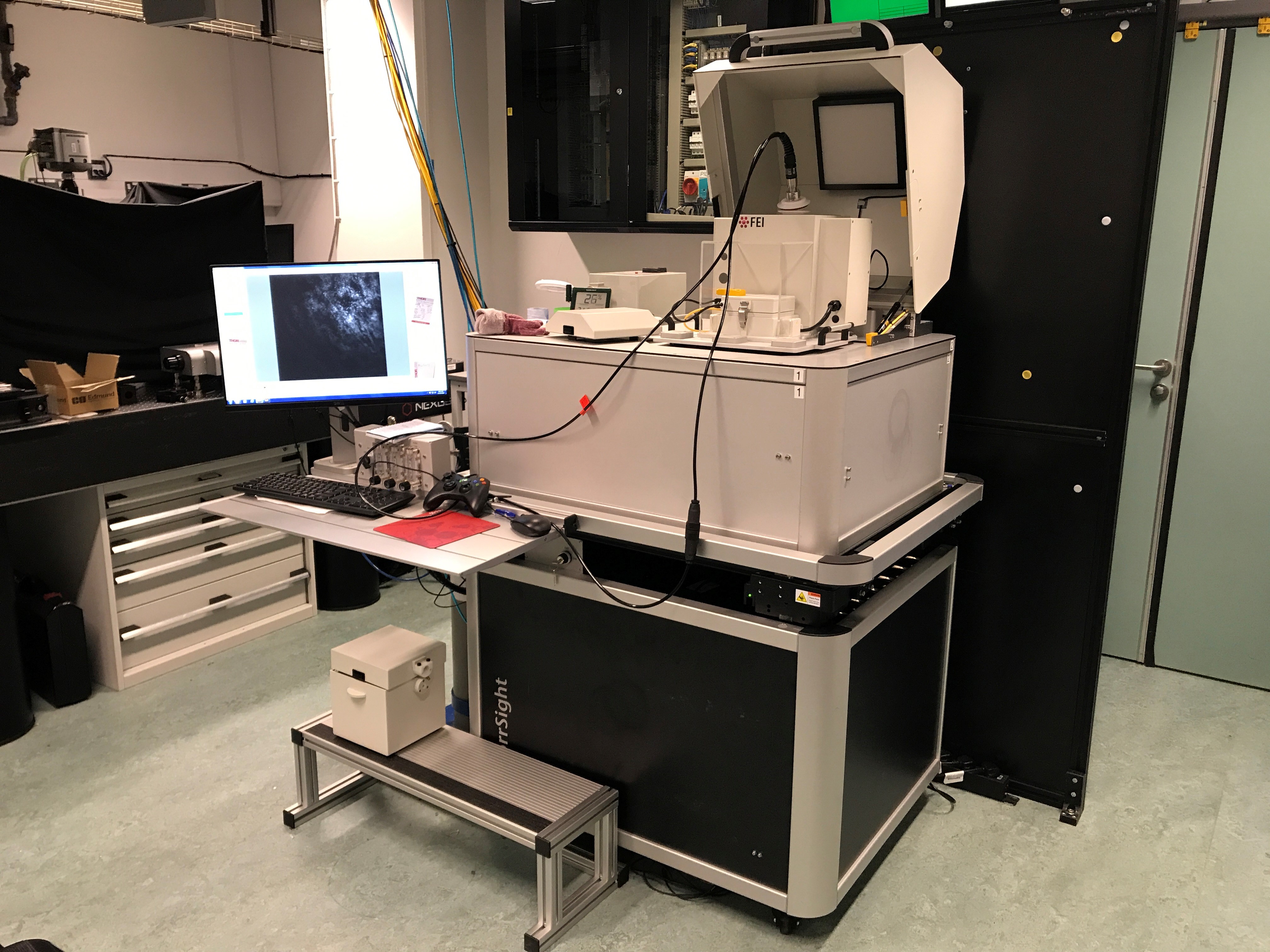
FEI CorrSight cryo-fluorescence microscope
This cryo-fluorescence microscope forms part of the correlative light and electron microscopy (CLEM) pipeline by fluorescently screening electron microscopy grids and discs prior to imaging on a transmission electron microscope. This instrument is user-friendly and offers mosaic mapping and auto-focussing algorithms for bulk throughput.
Lasers
| 405 nm, 488 nm, 561 nm, 642 nm @ suitable power densities for cryo-STORM.
|
Detector
| Hamamatsu Orca Flash V4.0.
|
Dichroics and filter sets
| We have combinations to allow for imaging most popular flourescent proteins and inorganic dyes.
Multi-colour imaging is achieved sequentially with fudicial registration.
|
Objectives
| 40x 0.9NA Zeiss
5x 0.16NA Zeiss
|
Suitable applications
| Fluorescence imaging of mammalian and bacterial cells.
STORM imaging of bacterial cells.
|
Resolution performance
| Diffraction limited optical imaging
STORM imaging: <30 nm localisation precisions on optimal samples
|
Temperature options
| -196°C and room temperature.
|
Leica cryo-ultramicrotome
This instrument provides easy sectioning of biological samples for use in SEM, TEM, and FM imaging. This is able to achieve slice thicknesses down to 100 nm.
FEI Vitrobot
We have access to an automated plunge-freezing system suitable for freezing EM grids, discs and our custom solid immersion lens assemblies. This instrument is designed to maximise the chances of achieving vitrous ice within your sample whilst providing reproduceable results through autonomy.
The Octopus cryo-imaging team
The multidisciplinery Octopus cryo-imaging team comprises of experts in microscopy, cell biology, engineering and data analysis.

Image taken February 2018. From left to right: Amy Moores, Lin Wang, Stephen Maughan, Benji Bateman, Laura Zanetti-Domingues, Sarah King, Alex McStea.